
Tutorial
1.Data access
Data retrieving in rSNPBase could be SNP-centric or gene-centric (Figure 1). SNP-centric data retrieving is appropriate to analyze results from genetic studies, especially the results of high-throughput studies, and then provide evidence for further functional studies to identify causal SNPs and shed light on underlying molecular mechanisms. Gene-centric searches are useful in candidate SNP selections that are based on genes of interest.Specifically, rSNPBase provides various search options (such as regulation type, tissue and developmental stage, and eQTL evidence) for gene-centric searches in 'Advanced search' module to facilitate data filtering.
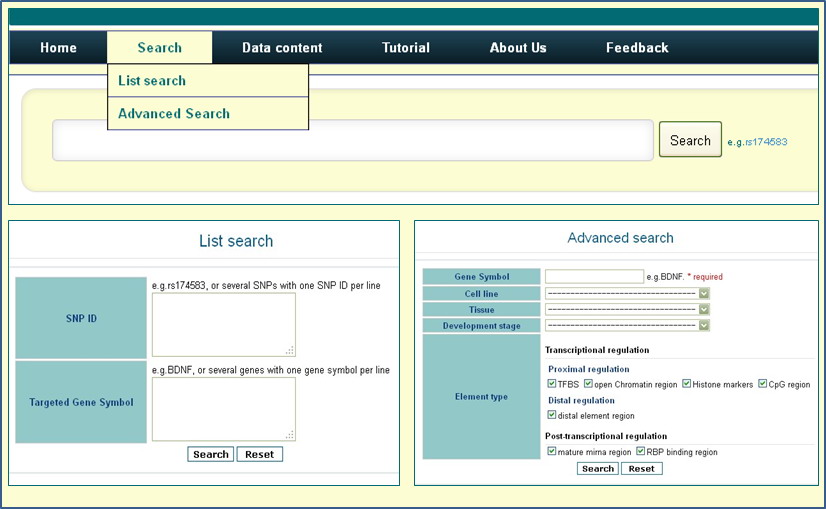
Figure 1 Data access of rSNPBase.
2.About search result
Search results of rSNPBase are summarized in a table on 'Search result' page (Figure 2), in which, the regulatory features of retrieved SNPs in four regulation manners are summarized as 'Yes' or 'No'.
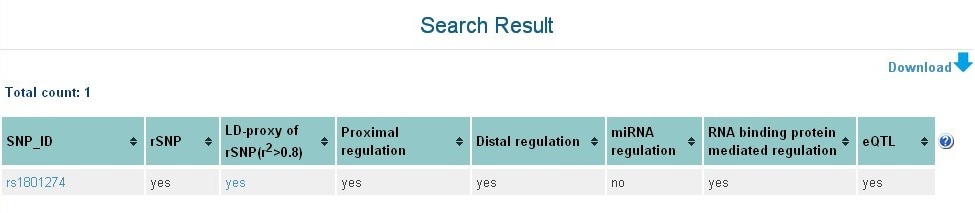
Figure 2 Search result page of rSNPBase.
Title of the table:- rSNP:rSNPBase identified regulatory SNPs.
- LD-proxy of rSNP(r2>0.8):SNP in strong LD (r2> 0.8) with rSNPs.
- Proximal regulation:SNP involved in proximal transcriptional regulation.
- Distal regulation:SNP involved in distal transcriptional regulation.
- miRNA regulation:SNP within mature miRNA.
- RNA binding protein mediated regulation:SNP involved in RNA binding protein-mediated post-transcriptional regulation
- eQTL:SNP with experimental eQTL evidence.
3.An example
Here, we use 110 SNPs from Jostinset al.(1) to make an example for SNP-centric date retrieval. Detailed description of these SNPs could be found in the GWAS catalog(2) (See in http://www.genome.gov/GWASStudySNPS.cfm?id=6987).
- To search a list of SNPs, we may choose the 'List search' module. Paste the SNP list into the textbox, submit it to the server and we will get the result page(Figure 3A).
- In the result page(http://rsnp.psych.ac.cn/result ), we could use the sort function of the table to choose SNPs of interest, i.e. sort the table with 'rSNP' or 'eQTL' and so on.(Figure 3B)
- Simply click on the SNP id and we will get a detailed report page, in which users can obtain a systematic view of each rSNP,its potential target genes and possible functions, and also extend the research field to its LD proxies.(Figure 3C)
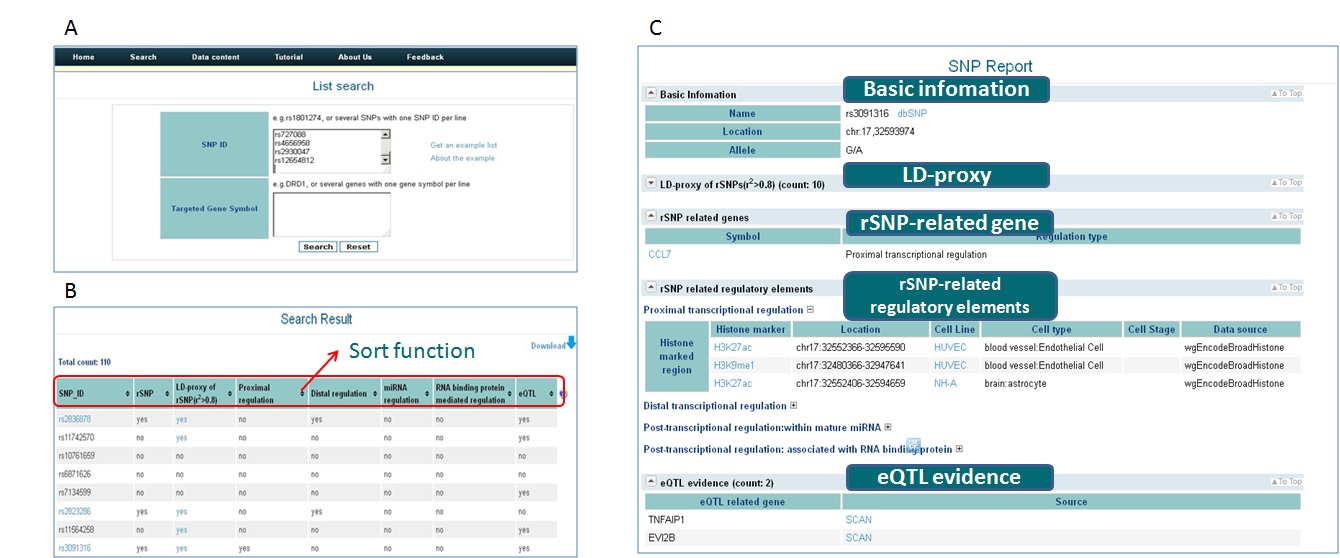
Figure 3 Data retrieval in rSNPBase.
Reference:
1. Jostins, L., Ripke, S., Weersma, R.K., Duerr, R.H., McGovern, D.P., Hui, K.Y., Lee, J.C., Schumm, L.P., Sharma, Y., Anderson, C.A. et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119-124. 2. Hindorff, L.A., Sethupathy, P., Junkins, H.A., Ramos, E.M., Mehta, J.P., Collins, F.S. and Manolio, T.A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America, 106, 9362-9367.